Malivhu
Malivhu: MAchine LearnIng for Virus classification and virus-HUman interaction prediction
About Malivhu
The outbreak of disease-causing agents is continuously threatening public health worldwide, particularly the viruses. Millions of people die due to the effects of illness resulting from virus infection, and over the years more aggressive epidemics are arising; the recent example being the ongoing COVID-19 pandemic. Host and pathogen are the life entities involved in such infectious diseases. The relationship between these two entities are crucial to the outcomes of infection and, therefore, host-pathogen interactions (HPI) have been studied to better understand the whole infection progression. HPIs are involved in every step of disease: from the initial pathogen transmission, through the activation of the pathogenic mechanism used to overcome (or hijack) host cell defenses, and to the pathogen becoming established and massively reproduce within the host system; all these processes involve the interaction of host and pathogen proteins. Identification of these protein-protein interactions (PPI) could help to unravel the disease pathways, provide methods to improve resistance, and ultimately accelerate the development of drugs and other therapeutics.
Knowing about PPIs is important for drug target discovery because we can enhance or disrupt these interactions to relieve diseases. To find about PPIs, there are high and low throughput methods, which are costly and there is a tradeoff between scalability and reliability due to high-throughput methods being able to output a lot of interactions, but many can be false positives. On the other hand, low-throughput methods are very reliable, but can detect a small number of interactions. This is where computational methods come in handy to predict if a pair of proteins interact, which could save a lot of resources and time.
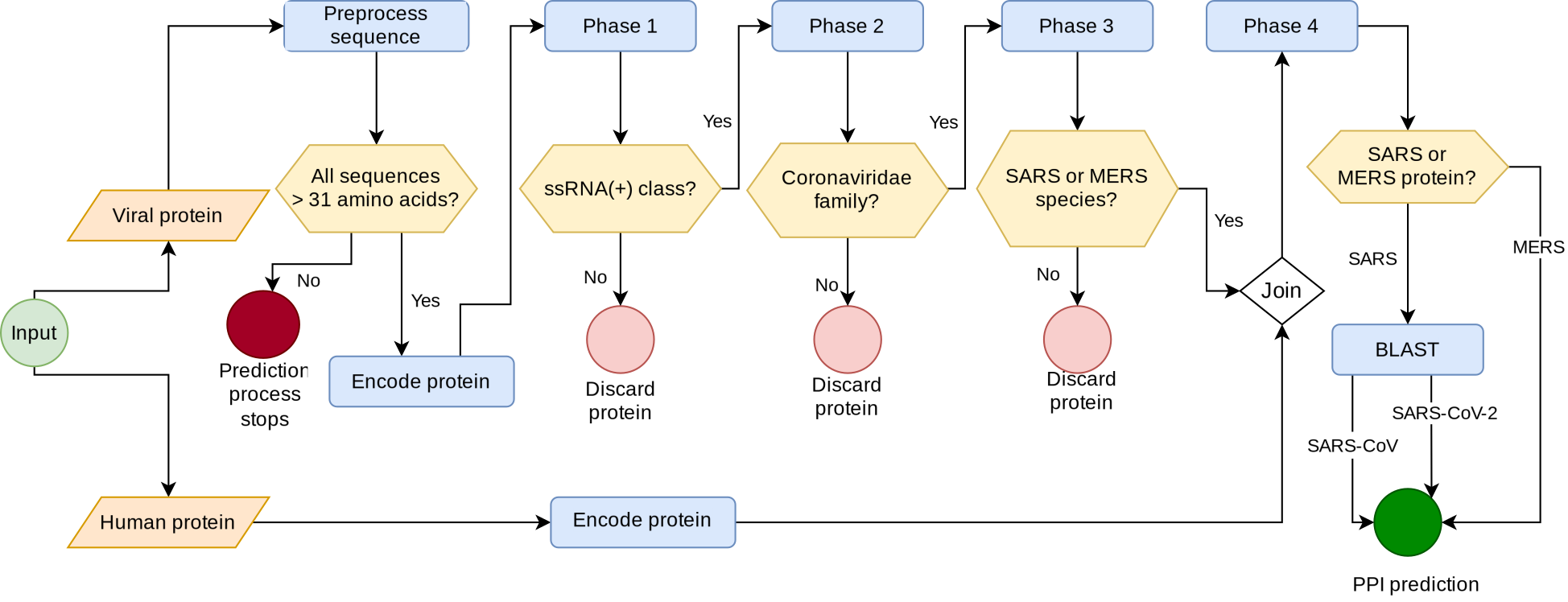
Malivhu lets the user input two protein sequences: the virus and the host proteins. In the background, it works in four phases: (I) it predicts whether the input sequence is an ssRNA(+) protein or not; (II) it predicts whether the protein comes from a coronavirus or not; (III) it predicts whether the protein comes from MERS or SARS or other species; and (IV) runs a BLAST for all SARS proteins to find if they belong to SARS-CoV or SARS-CoV-2, and then it predicts whether both proteins interact or not.