HuCoVaria
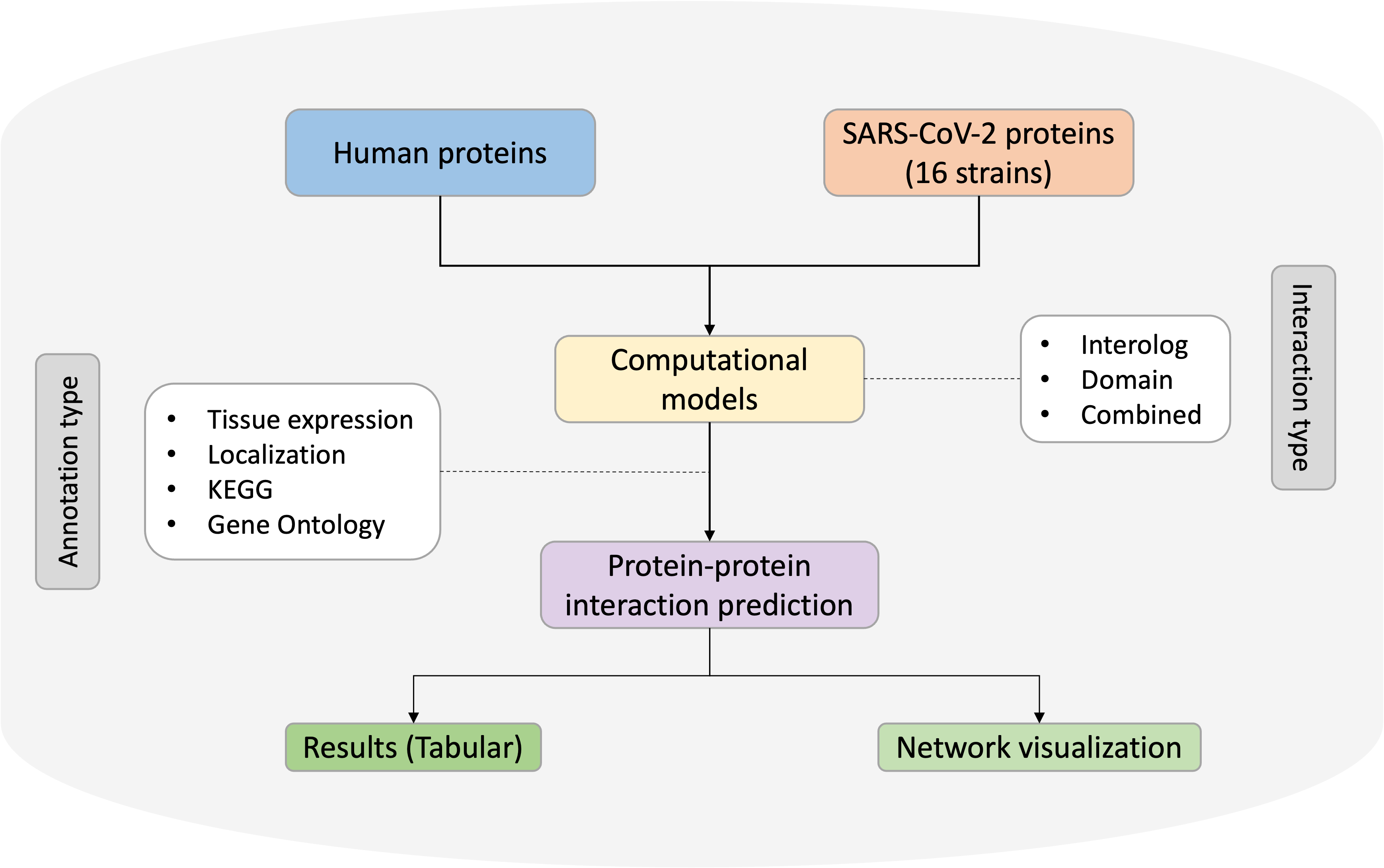
The SARS-CoV-2 virus continues to mutate significantly, potentially altering its transmissibility and pathogenicity. Therefore, understanding the protein-protein interactions between viral and human proteins can shed light on the shared molecular mechanisms and biological pathways involved across various viral strains.
The interactions inside HuCoVarIA are categorized into three groups: interologs, predicted using BLAST, domains, predicted using HMMER, and machine learning-based, using a neural network. There is also an option to combine the interactions from interologs and domains. In total, 171,676 interactions have been identified across 16 strains, including the original reference strain, using interolog and domain methods, while the machine learning-based method predicted 4,505,779 interactions. These interactions have been further characterized using KEGG for pathway enrichment, Gene Ontology for functional enrichment, subcellular localization analysis, and tissue expression profiling. Users can navigate this database with any of the above features. The comparative interactomes among these strains can be visualized in an enriched manner, and user-customized networks can be displayed, thus providing an enhanced visualization experience.
We believe that the comparison of different strains in HuCoVarIA has potential applications in the development of pan-viral vaccine and drug development.
For any problems, issues or questions that arise from using this webtool, e-mail bioinfo@kaabil.net.