RSLpred-2.0 Tutorial
Introduction
Help section page of RSLpred-2.0, here you will find a step by step guide to submit and display the result of an analysis, as well as the different options regarding prediction on RSLpred-2.0 and what each parameter means. If you have any questions that are not covered in this page please refer to FAQ page or send an email to info@kaabil.net. For this tutorial we will use the Demo data, so if you want to replicate the results obtained in this tutorial just click in the load Demo FASTA link.
This website is free and open to all users and there is no login requirement.Data input
RSLpred-2.0 supports FASTA format. You can either upload a file or paste your sequences in the text area below or provide NCBI/UniProt accession in the text box below. You can upload amino acid sequences after selecting appropriate query sequence type below.

Prediction Models
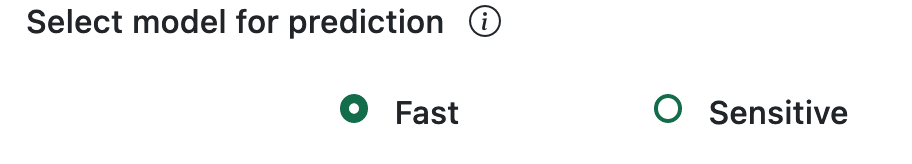
There are two prediction strategy available in RSLpred-2.0 [Fast, Sensitive]. There is an icon with general information about the tool and a brief explanation on how it works.
The 'Fast' approach model is designed for swift predictions. In its fast mode, the model utilizes a DPCP feature, a composite of Dipeptide composition (400), amino acid pair count (400), and Schneider-Wrede values for each amino acid pair (400). The resulting DPCP feature vector has a total size of 1200. The efficiency of this approach is attributed to its smaller vector size, facilitating rapid processing. This makes the 'Fast' approach particularly beneficial for annotating a large volume of proteins with speed and accuracy.
The 'Sensitive' approach model excels in delivering heightened sensitivity in predictions, albeit with a trade-off of increased computation time. In its sensitive mode, the model employs the TPC (tripeptide composition) feature, generating an extensive 8000-length vector. The extended computation time is a result of the larger vector size, making this approach particularly sensitive. However, the 'Sensitive' approach proves valuable for annotating a smaller number of proteins with a focus on high-quality predictions.
Prediction Phases
RSLpred-2.0 runs four levels for Oryza sativa Subcellular Localization prediction. Users can select any level for prediction and their preceeding levels will be automatically selected. All levels will run by default. There is an icon with general information about the tool and a brief explanation on how it works.
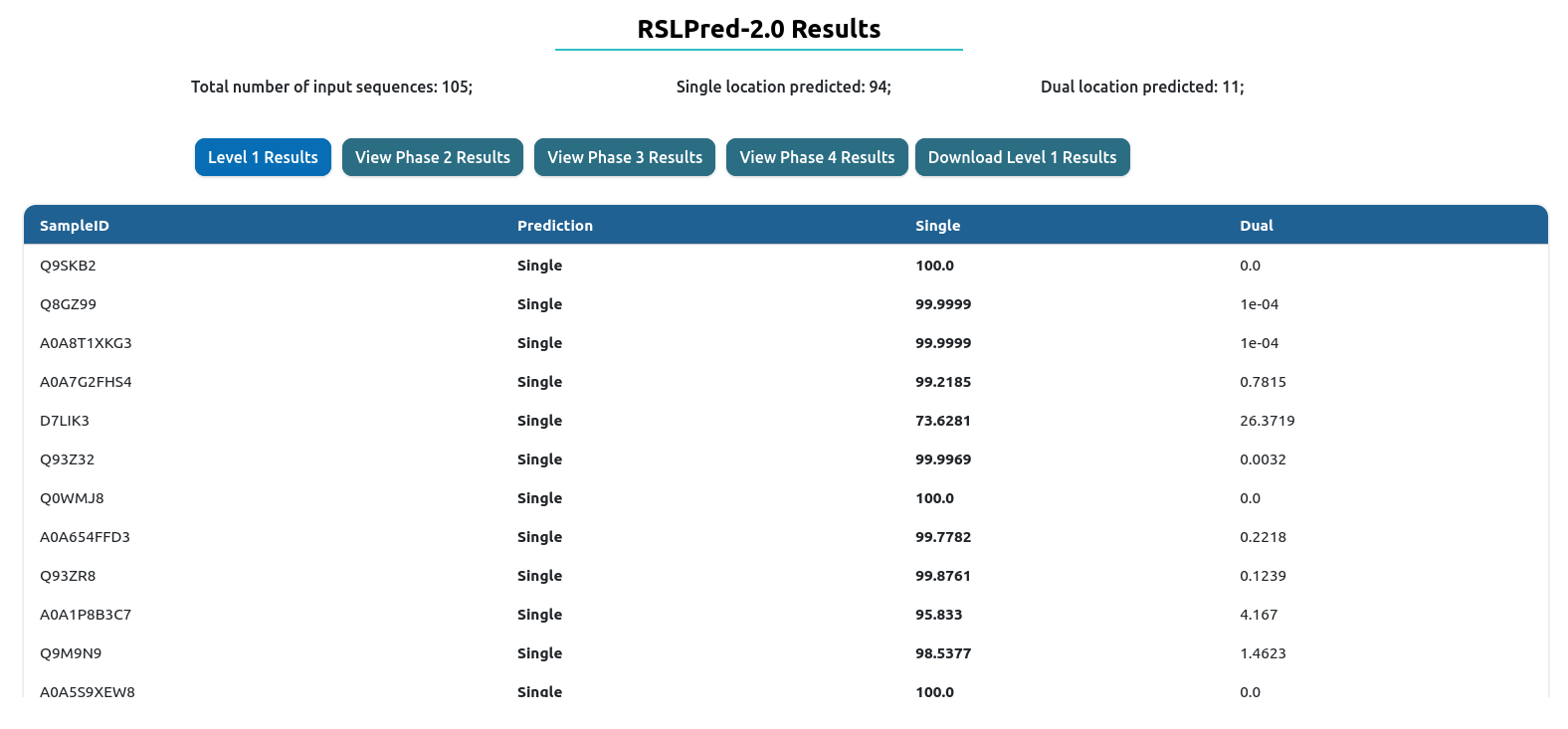
Level I: This is first level where a query sequence is being predicted as single localization or dual localization

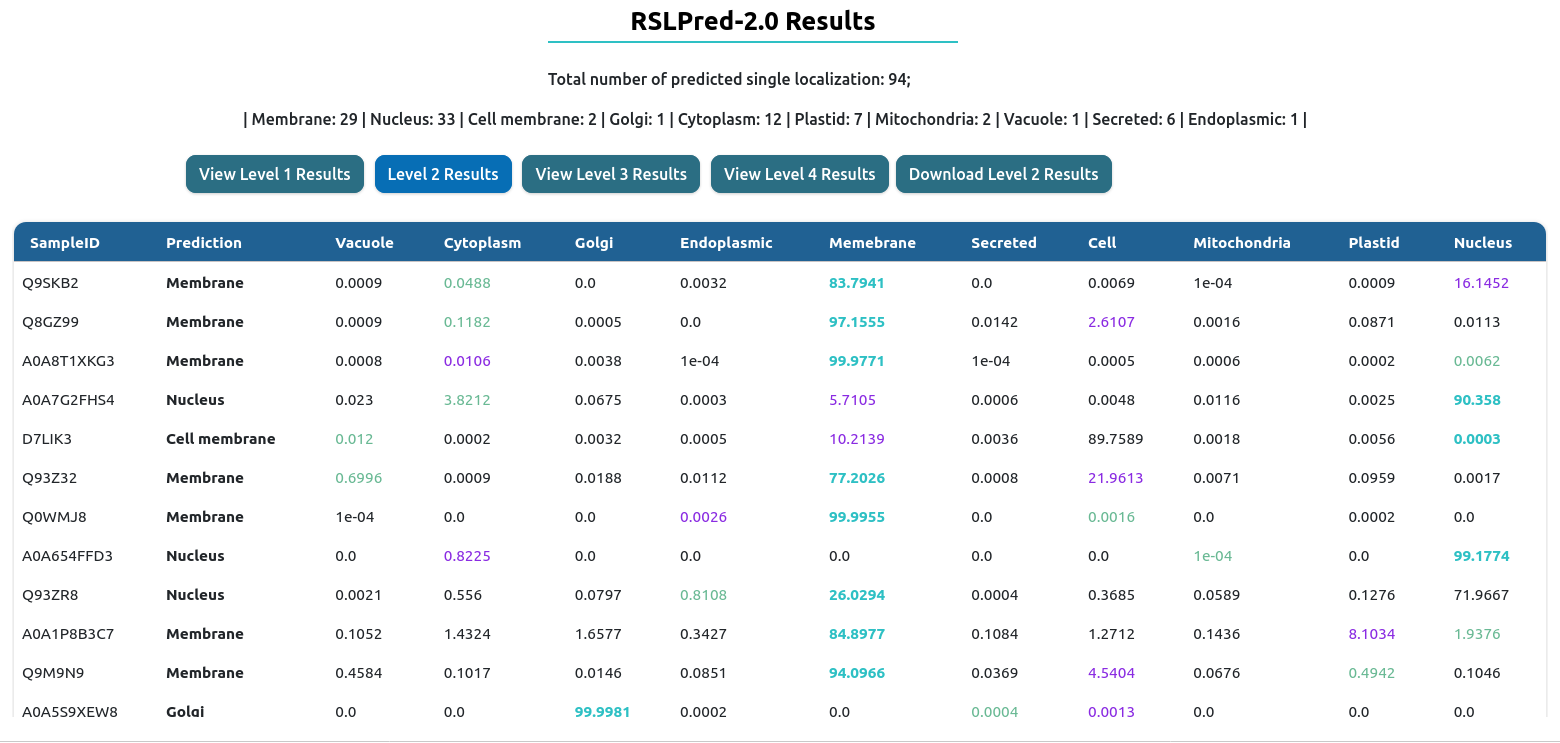
Level II: This level will run leve1 first followed by level2 where a query sequence is first predicted as single localization or dual localization followed by classifying single localization into 10 classes. 'Vacuole', 'Cytoplasm', 'Golgi', 'Endoplasmic', 'Memebrane', 'Secreted', 'Cell', 'Mitochondria', 'Plastid','Nucleus'.

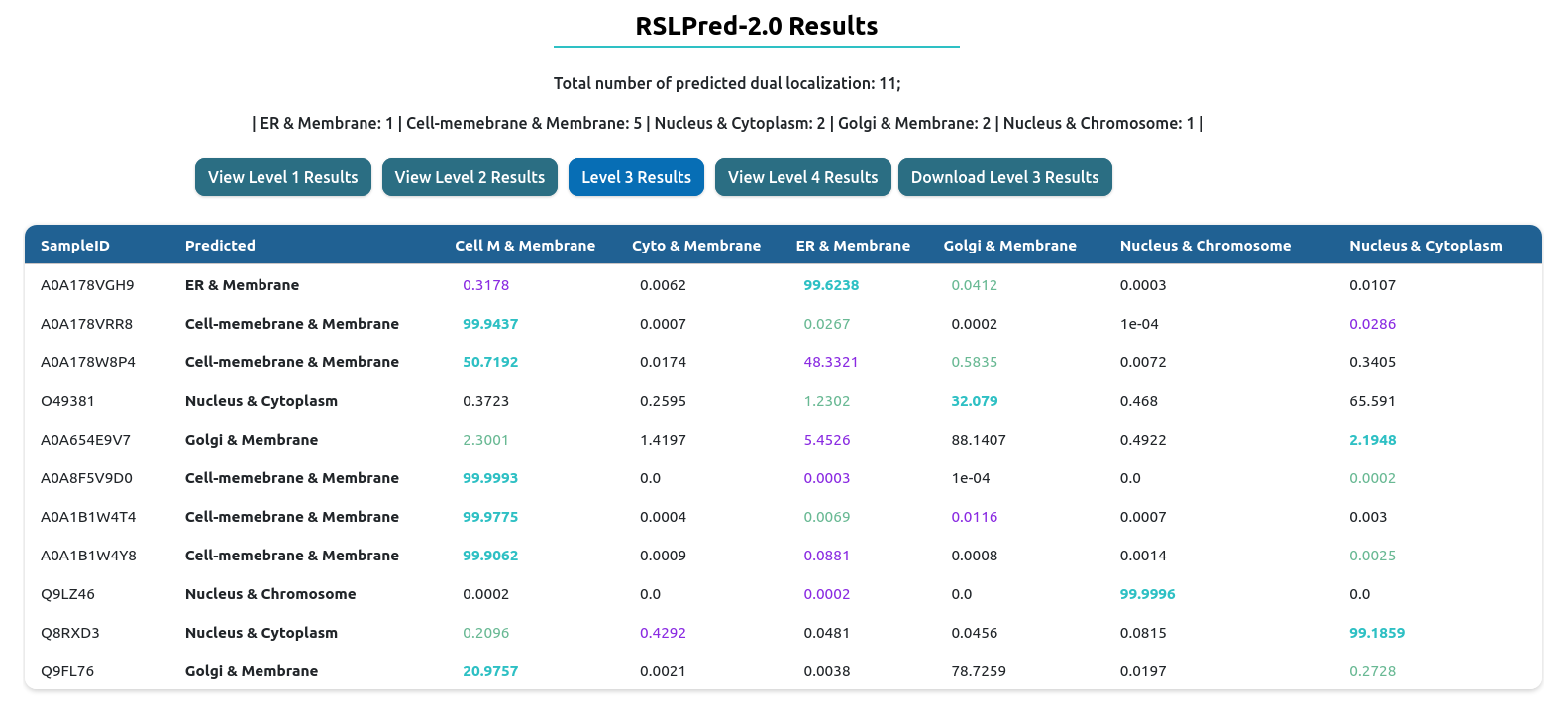
Level III: This level will execute level1 and level2 followed by classifying dual localization in 6 classes. 'Cell-memebrane & Membrane', 'Cytoplasm & Membrane','Endoplasmic reticulum & Membrane','Golgi & Membrane','Nucleus & Chromosome','Nucleus & Cytoplasm'.

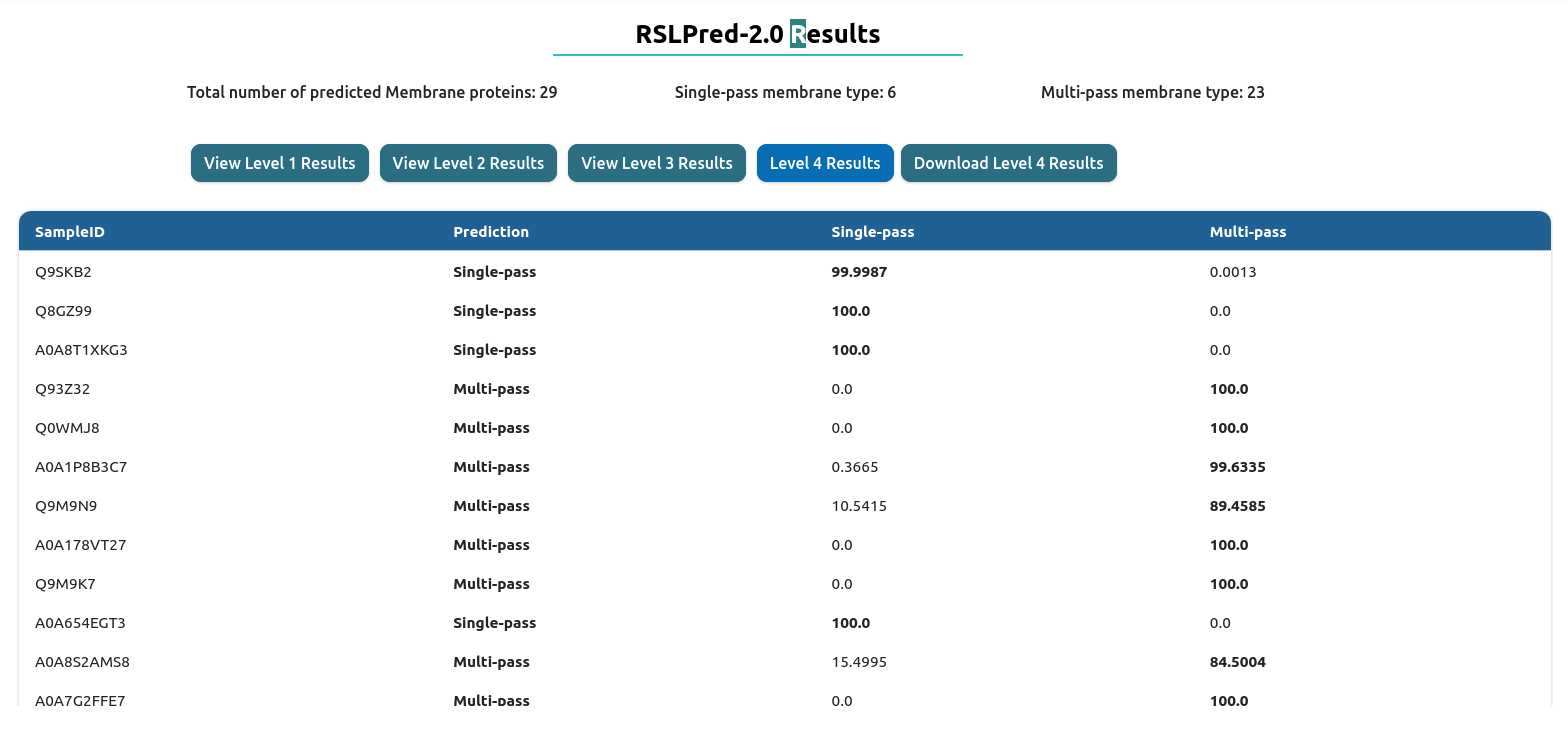
level IV: This phase will execute level1, level2 and level3 followed by classifying Memebrane predicted in level2 into single-pass or multi-pass.

Run Prediction
Run Prediction section contains an input box in which you can provide an email to receive a link to your results when they are done. You can reset the form. Run prediction button will execute the prediction on RSLpred-2.0 server.
Output Example
The result table will change depending of the prediction level used. On the top, you will have download results button, which download the comprehensive table result in tab-delimited format.
Browser Compatibility
RSLpred-2.0 have been tested in the following setups.
| OS | Version | Chrome | Firefox | Safari | Edge |
|---|---|---|---|---|---|
| Linux | Ubuntu 22.04 | 108.0.5359.71 | 112.0.2 | n/a | 113.0.1774.35 |
| MacOS | Ventura 13.3.1 (a) | 108.0.5359.71 | 112.0.2 | 16.4 | 113.0.1774.35 |
| Windows | 10 | 108.0.5359.71 | 112.0.2 | not tested | 113.0.1774.35 |